Contributing to diverse research opportunities
Explore project documentation below.
Statement of Purpose
My research journey began at the miRcore Computational Biology Camp in 2022. That one week gave me access to programs that connected me with other high schoolers equally fascinated by gene networks, disease pathways, data science, and eventually machine learning. With my expansion to other research projects, I found more than just collaborators—I found a scientific family. These moments solidified my dream: to pursue medicine with a foundation deeply rooted in research, and I’m excited for the opportunity to conduct novel research at Penn.

High School Projects (miRcore and BWSI)
This study utilized the GSE190847 dataset, which contains B-cell transcriptome profiles, to investigate gene expression differences associated with Multiple Sclerosis (MS). By applying analytical tools such as GEO2R for differential gene expression, STRING for protein-protein interaction mapping, and KEGG for pathway enrichment analysis, the research identified key genes and biological pathways significantly involved in the pathogenesis of MS. These findings contribute to a deeper understanding of the molecular mechanisms driving the disease and may inform future therapeutic strategies.

NIH Clinical Trial: MONACO
We developed a machine learning model to predict patient mortality outcomes using clinical tabular data from the Uterine Corpus Endometrial Carcinoma (UCEC) dataset within the TCGA PanCancer Atlas. Leveraging the rich clinical features provided in the dataset, we designed and implemented a neural network architecture to capture complex, non-linear relationships between variables and improve predictive performance. This approach allowed us to build a robust and data-driven model capable of identifying patterns associated with patient survival, contributing valuable insights to the field of cancer prognosis and personalized medicine.
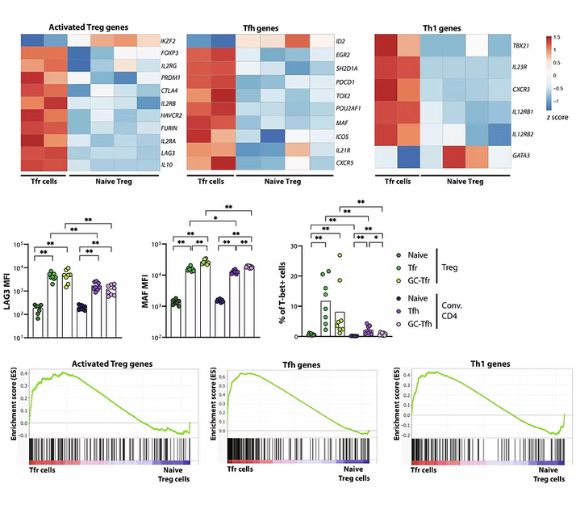
Immunology Translational Research: Locci Lab
We examined dataset GSE219278, which includes genetic variant data related to Frontotemporal Dementia (FTD), using a range of statistical and bioinformatic tools to uncover underlying biological mechanisms. Through comprehensive analysis, including differential gene expression and pathway enrichment assessments, we identified a significant inhibition of the oxidative phosphorylation pathway in patients with Multiple Sclerosis. This discovery suggests a potential link between disrupted mitochondrial energy metabolism and the pathophysiology of MS, ultimately pointing out possible targets.
Research Next Steps
Due to the limitations of my location, many of my research experiences have been conducted virtually, relying primarily on publicly available datasets and the analytical techniques I’ve learned. While these opportunities have been incredibly valuable in building my foundational skills, they have also highlighted the constraints of remote research. To me, college represents a gateway to transformative resources—labs, mentorship, and hands-on experiences—that will allow me to engage in research on a much deeper level. Rather than working solely with existing mRNA data from sources like the NIH, I look forward to generating and analyzing my own, contributing meaningfully to the scientific process from start to finish.
